
Download PDF
Translations (PDF)
Visit r-pkgs.org to learn more about writing and publishing packages for R.
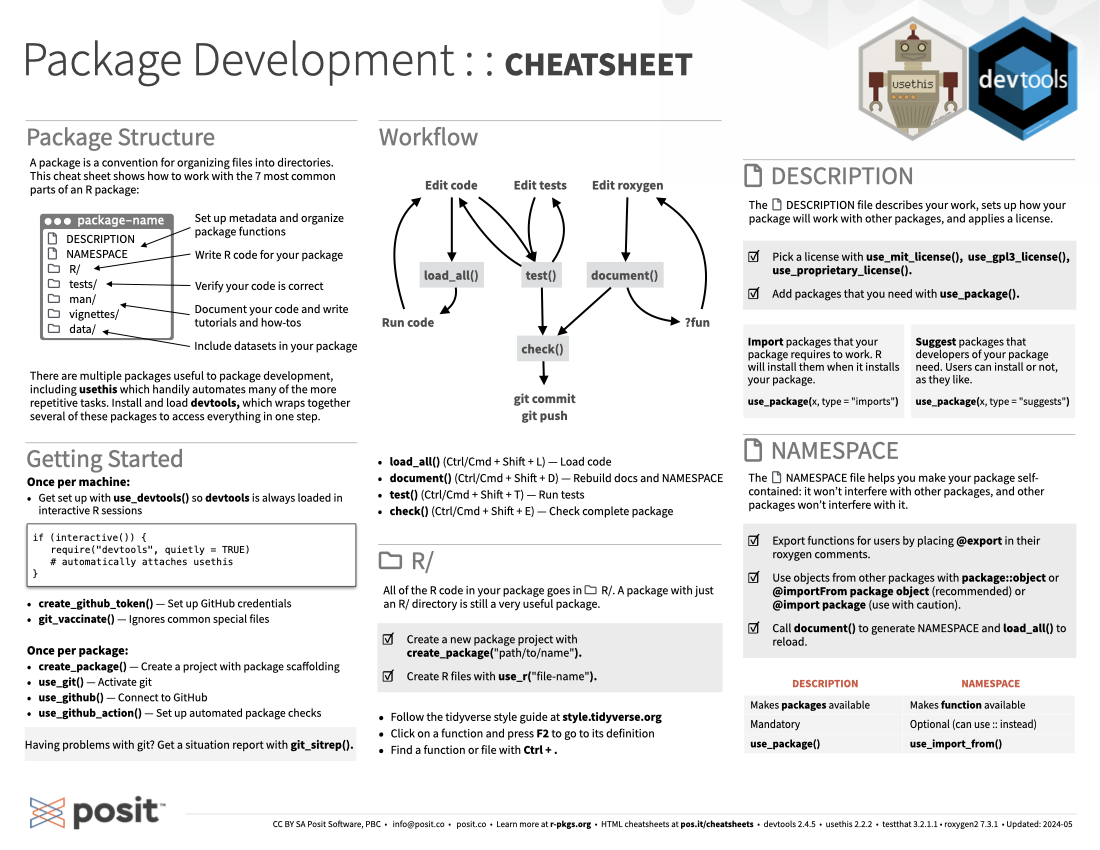
A package is a convention for organizing files into directories, and creates a shareable, installable collection of functions, sample data, and documentation. This cheatsheet shows you how to work with the 7 most common parts of an R package:
There are multiple packages useful to package development, including usethis which handily automates many of the more repetitive tasks. Load and install devtools which wraps together several of these packages to access everything in one step.
Once per machine:
Get set up with use_devtools() so devtools is always loaded in interactive R sessions.
create_github_token(): Set up GitHub credentials.
git_vaccinate(): Ignores common special files.
Once per package:
create_package(): Create a project with package scaffolding.
use_git(): Activate git.
use_github(): Connect to GitHub.
use_github_action(): Set up automated package checks.
Having problems with git? Get a situation report with git_sitrep().
load_all() (Ctrl/Cmd + Shift + L): Load codetest() (Ctrl/Cmd + Shift + T): Run testsdocument() (Ctrl/Cmd + Shift + D): Rebuild docs and NAMESPACEcheck() (Ctrl/Cmd + Shift + E): Check complete packageAll of the R code in your package goes in R/. A package with just an R/ directory is still a very useful package.
Create a new package project with create_package("path/to/name").
Create R files with use_r("file-name").
Follow the tidyverse style guide at style.tidyverse.org
Put your cursor on a function and press F2 to go to its definition
Find a function or file with the keyboard shortcut Ctrl+.
The DESCRIPTION file describes your package, sets up how your package will work with other packages, and applies a license.
Pick a license with use_mit_license(), use_gpl3_license(), use_proprietary_license().
Add packages that you need with use_package().
Import packages that your package requires to work. R will install them when it installs your package. Add with use_package(pkgname, type = "imports")
Suggest packages that developers of your package need. Users can install or not, as they like. Add with use_package(pkgname, type = "suggests")
The NAMESPACE file helps you make your packages self-contained: it won’t interfere with other packages, and other packages won’t interfere with it.
Export functions for users by placing @export in their roxygen comments.
Use objects from other packages with package::object or @importFrom package object (recommended) or @import package (use with caution).
Call document() to generate NAMESPACE and load_all() to reload.
| DESCRIPTION | NAMESPACE |
|---|---|
| Makes packages available | Makes functions available |
| Mandatory | Optional (can use :: instead) |
use_package() |
use_import_from() |
The documentation will become the help pages in your package.
Document each function with a roxygen block above its definition in R/. In RStudio, Code > Insert Roxygen Skeleton (Keyboard shortcut: Mac Shift+Option+Cmd+R, Windows/Linux Shift+Alt+Ctrl+R) helps.
Document each data set with an roxygen block above the name of the data set in quotes.
Document the package with use_package_doc().
Build documentation in man/ from Roxygen blocks with document().
The roxygen2 package lets you write documentation inline in your .R files with shorthand syntax.
Add roxygen documentation as comments beginning with #'.
Place an roxygen @ tag (right) after #' to supply a specific section of documentation.
Untagged paragraphs will be used to generate a title, description, and details section (in that order).
@examples@export@param@returnsAlso:
@description@examplesif@family@inheritParams@rdname@seealsouse_vignette().use_article().use_pkgdown_github_pages() to set up pkgdown and configure an automated workflow using GitHub Actions and Pages.use_pkgdown() to configure pkgdown. Then build locally with pkgdown::build_site().use_readme_rmd() and use_news_md().use_test().test_that() and expect_().test() and run tests for current file with test_active_file().test_coverage() and see coverage of current file with test_coverage_active_file().| Expect statement | Tests |
|---|---|
expect_equal() |
Is equal? (within numerical tolerance) |
expect_error() |
Throws specified error? |
expect_snapshot() |
Output is unchanged? |
data-raw/ with use_data_raw().data/ with use_data().The contents of a package can be stored on disk as a:
Packages exist in those states locally or remotely, e.g. on CRAN or on GitHub.
From those states, a package can be installed into an R library and then loaded into memory for use during an R session.
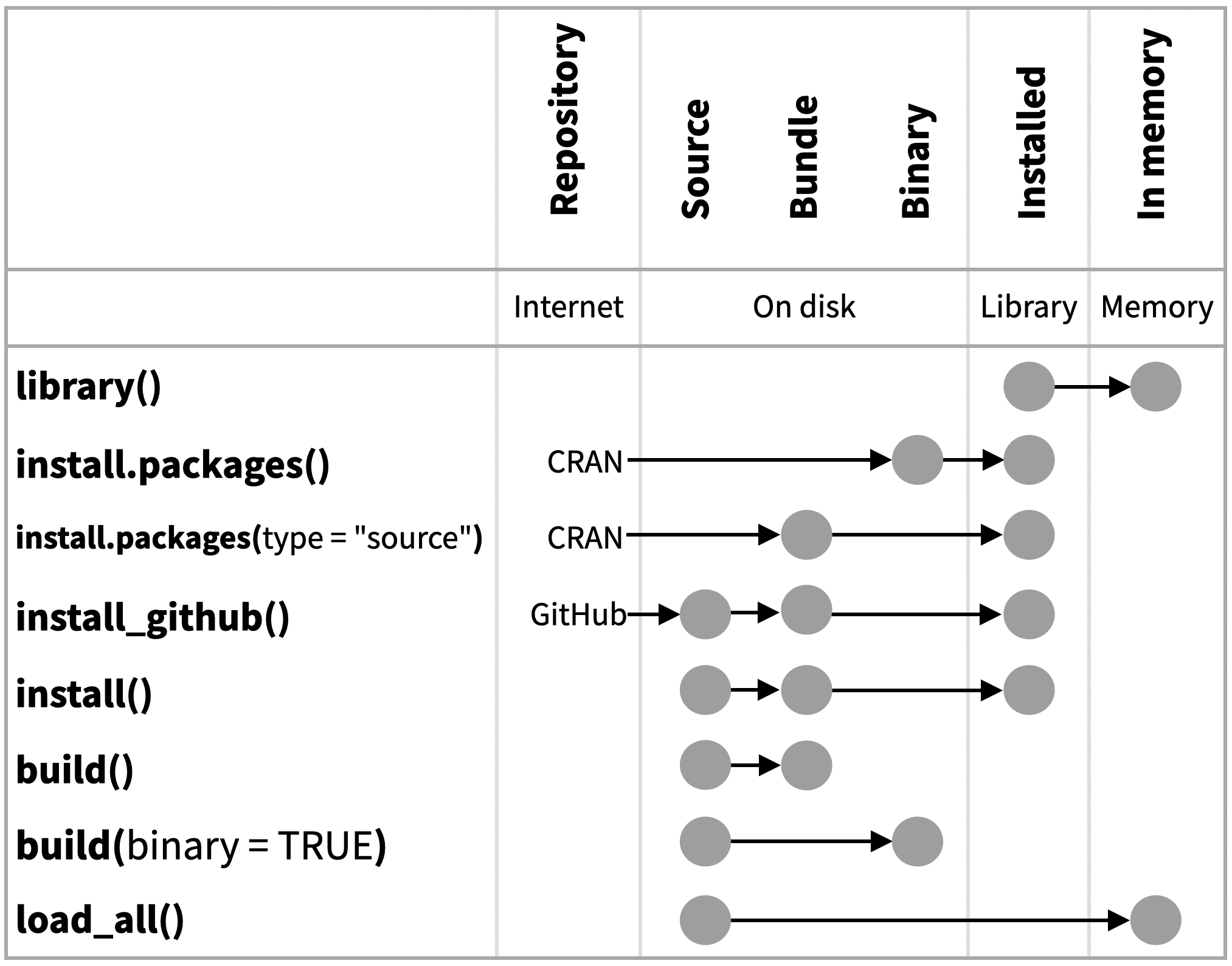
Use the functions below to move between these states:
library(): Installed in Library to loaded in Memory.install.packages(): Binary from CRAN repository to installed in Library.install.packages(type = "source"): Source code from CRAN repository to Bundle, to installed in Library.install_github(): Source code from GitHub repository to Bundle to installed in Library.install(): Local source code to bundle to installed in Library.build(): Local source to Bundle.build(binary = TRUE): Local source to Binary.load_all(): Local source to loaded in Memory.CC BY SA Posit Software, PBC • info@posit.co • posit.co
Learn more at r-pkgs.org.
Updated: 2024-05.
[1] '2.4.5'[1] '2.2.3'[1] '3.2.1.1'[1] '7.3.1'