Typically, models in R exist in memory and can be saved as
.rds files. However, some models store information in
locations that cannot be saved using save() or
saveRDS() directly. The goal of bundle is to provide a
common interface to capture this information, situate it within a
portable object, and restore it for use in new settings.
This vignette walks through how to prepare a statistical model for saving to demonstrate the benefits of using bundle.
In addition to the package itself, we’ll load the keras and xgboost packages to fit some example models, and the callr package to generate fresh R sessions to test our models inside of.
library(keras)
#> The keras package is deprecated. Use the keras3 package instead.
library(xgboost)
library(callr)Saving things is hard
As an example, let’s fit a model with the keras package, building a
neural network that models miles per gallon using the rest of the
variables in the built-in mtcars dataset.
cars <- mtcars |>
as.matrix() |>
scale()
x_train <- cars[1:25, 2:ncol(cars)]
y_train <- cars[1:25, 1]
x_test <- cars[26:32, 2:ncol(cars)]
y_test <- cars[26:32, 1]
keras_fit <-
keras_model_sequential() |>
layer_dense(units = 1, input_shape = ncol(x_train), activation = 'linear') |>
compile(
loss = 'mean_squared_error',
optimizer = optimizer_adam(learning_rate = .01)
)
keras_fit |>
fit(
x = x_train, y = y_train,
epochs = 100, batch_size = 1,
verbose = 0
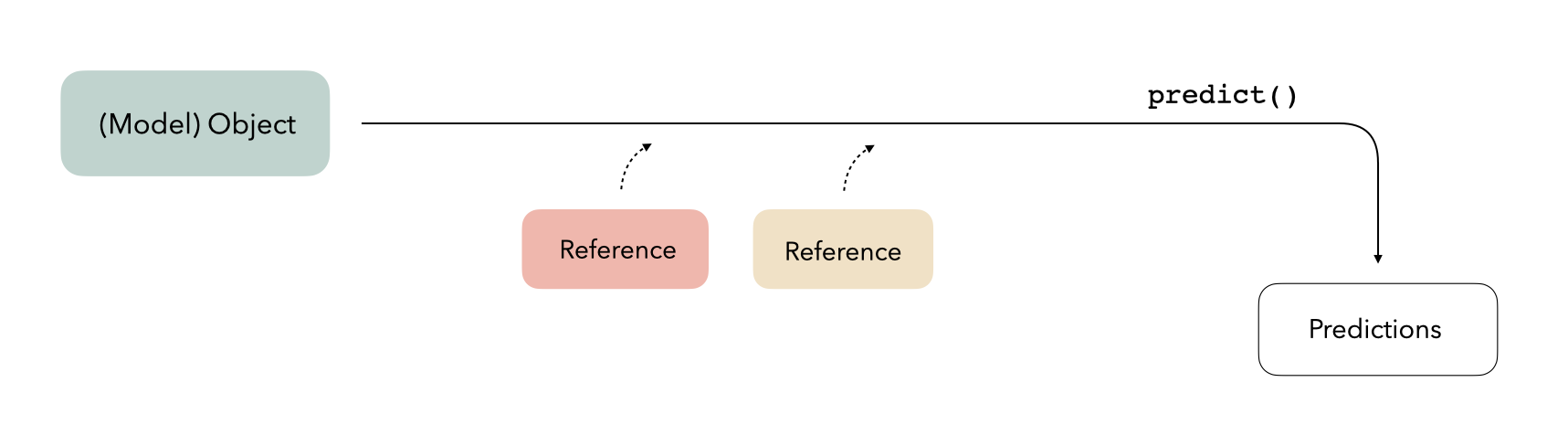
)Easy peasy! Now, given that this model is trained, we assume that it’s ready to go to predict on new data. Our mental map might look something like this:

We pass a model object to the predict() function, along
with some new data to predict on, and get predictions back. Let’s try
that out:
predict(keras_fit, x_test)
#> 1/1 - 0s - 44ms/epoch - 44ms/step
#> [,1]
#> [1,] 1.4218841
#> [2,] 1.6472901
#> [3,] 1.0359328
#> [4,] 0.6768160
#> [5,] -0.5854317
#> [6,] -1.5305036
#> [7,] 0.9310328Perfect.
If we’re satisfied with this model and think it provides some valuable insights, we might want to deploy it somewhere—maybe as a REST API or as a Shiny app—so that others can make use of it.
The callr package will be helpful for emulating this kind of situation. The package allows us to start up a fresh R session and pass a few objects in.
We’ll just make use of two of the arguments to the function
r():
-
func: A function that, given a model object and some new data, will generate predictions, and -
args: A named list, giving the arguments to the above function.
As an example:
So, our approach might be:
- save our model object
- start up a new R session
- load the model object into the new session
- predict on new data with the loaded model object
First, saving our model object to a file:
Now, starting up a fresh R session and predicting on new data:
r(
function(temp_file, new_data) {
library(keras)
model_object <- readRDS(file = temp_file)
predict(model_object, new_data)
},
args = list(
temp_file = temp_file,
new_data = x_test
)
)
#> Error: ! in callr subprocess.
#> Caused by error in `do.call(object$predict, args)`:
#> ! 'what' must be a function or character stringOof. Hm.
After a bit of poking around in keras’ documentation, you might come
across the keras vignette “Saving and serializing models” at
vignette("saving_serializing", package = "keras"). That
vignette points us to several functions in the keras package
that will allow us to save our model fit in a way that allows it to
predict in a new session.
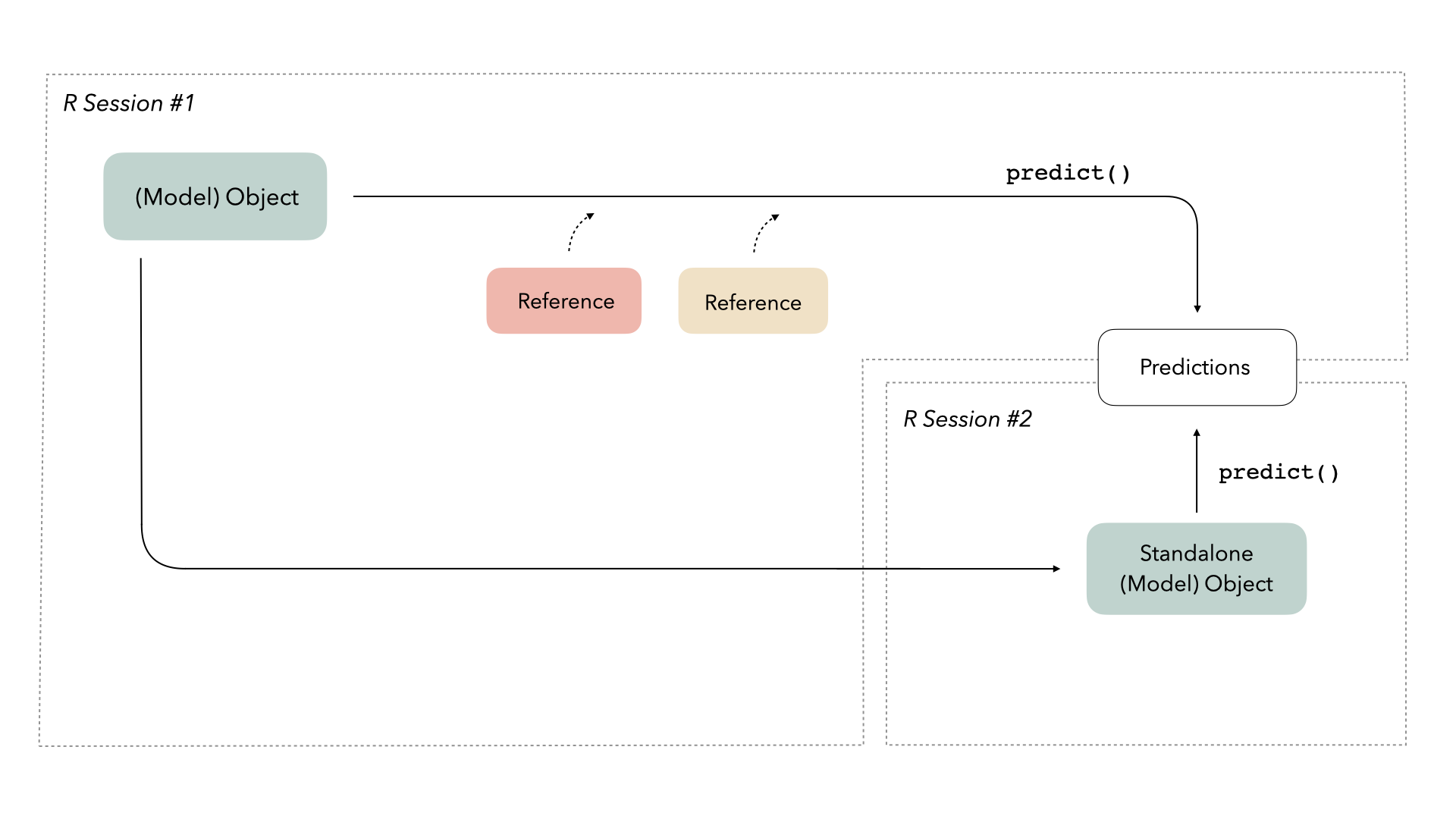
Given this new understanding, we can update our mental map a bit. Some objects require extra information when they’re loaded into new environments in order to do their thing. In this case, this keras model object needs access to additional references in order to predict on new data.
In computer science, these bits of “extra information” are called references. Those references need to persist—or be restored—in new environments in order for the objects that reference them to work well.
These kinds of custom methods to save objects, like the ones that keras provide, are often referred to as native serialization. Methods for native serialization know which references need to be brought along in order for an object to effectively do its thing in a new environment.
Let’s make use of native serialization, then!
Native serialization, and where it falls short
keras’ vignette is really informative in telling us what we ought to
do from here; if we save the model with the native serialization rather
than saveRDS, we’ll be good to go.
Saving our model object with their methods:
temp_dir <- tempdir()
save_model_tf(keras_fit, filepath = temp_dir)Now, starting up a fresh R session and predicting on new data:
r(
function(temp_dir, new_data) {
library(keras)
model_object <- load_model_tf(filepath = temp_dir)
predict(model_object, new_data)
},
args = list(
temp_dir = temp_dir,
new_data = x_test
)
)
#> [,1]
#> [1,] 1.4218841
#> [2,] 1.6472901
#> [3,] 1.0359328
#> [4,] 0.6768160
#> [5,] -0.5854317
#> [6,] -1.5305036
#> [7,] 0.9310328Awesome! Making use of their methods, we were able to effectively save our model, load it in a new R session, and predict on new data.
Now let’s consider a new scenario—I’ve heard that xgboost models are super performant, and want to try to productionize those, too. How would we do that?
Based on our workflow just now, we could try to just save it with
saveRDS and see if we get an informative error somewhere
along the way to predicting in a new R session. Or, maybe a better
approach would be to read through their documentation and see if we can
find anything related to serialization.
We’ve done the work of figuring that out, and it turns out the
interface is a little bit different. You’ll need to make sure the
params object persists across sessions, but
saveRDS will work by itself if… ah, I’ll stop myself
there.
What if we could just use the same function for any R object, and it would just work?
Using bundle
bundle provides a consistent interface to prepare R model objects to
be saved and re-loaded. The package provides two functions,
bundle() and unbundle(), that take care of all
of the minutae of preparing to save and load R objects effectively.
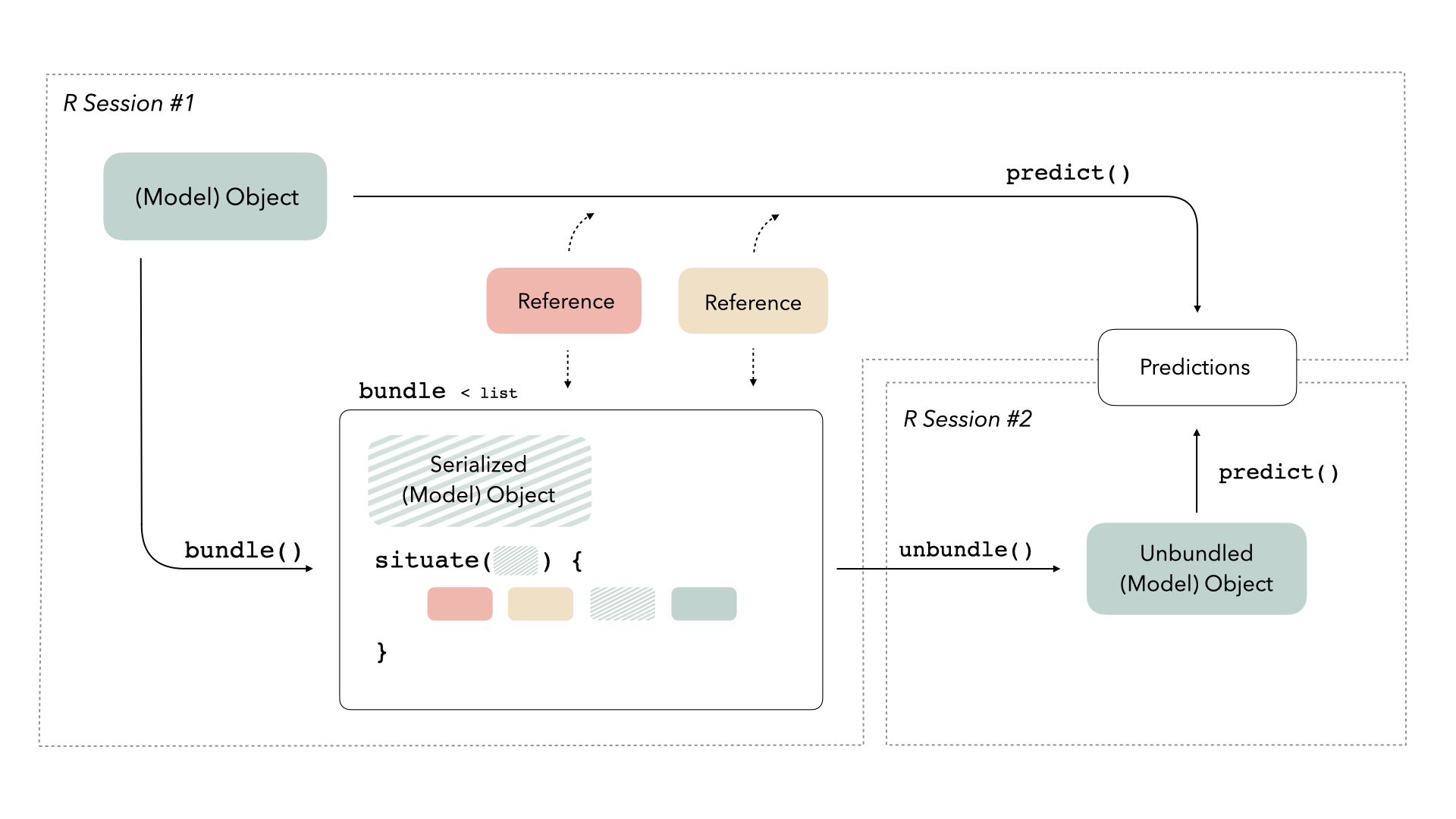
Bundles are just lists with two elements:
-
object: Theobjectelement of a bundle is the serialized version of the original model object. In the simplest situations in modeling, this object is just the output of a native serialization function likesave_model_tf()that we used earlier. -
situate(): Thesituate()element of a bundle is a function that situates theobjectelement in its new environment. It takes in theobjectelement as input, but also “freezes” reference that existed when the original object was created.
When unbundle() is called on a bundle object, the
situate() element of the bundle will re-load the
object element and restore needed references in the new
environment. Thus, the output of unbundle() is ready to go
for prediction wherever it is called.
To be a bit more concrete, lets return to the keras example. Bundling the model fit:
keras_bundle <- bundle(keras_fit)Now, starting up a fresh R session and predicting on new data:
r(
function(model_bundle, new_data) {
library(bundle)
model_object <- unbundle(model_bundle)
predict(model_object, new_data)
},
args = list(
model_bundle = keras_bundle,
new_data = x_test
)
)
#> [,1]
#> [1,] 1.4218841
#> [2,] 1.6472901
#> [3,] 1.0359328
#> [4,] 0.6768160
#> [5,] -0.5854317
#> [6,] -1.5305036
#> [7,] 0.9310328Huzzah!
The best part is, if you wanted to do the same thing for an xgboost object, you could use the same code!
First, fitting a quick xgboost model:
xgb_fit <-
xgboost(
data = x_train,
label = y_train,
nrounds = 5
)
#> Warning in throw_err_or_depr_msg("Parameter '", match_old, "' has been
#> renamed to '", : Parameter 'data' has been renamed to 'x'. This
#> warning will become an error in a future version.
#> Warning in throw_err_or_depr_msg("Parameter '", match_old, "' has been
#> renamed to '", : Parameter 'label' has been renamed to 'y'. This
#> warning will become an error in a future version.Now, bundling it:
xgb_bundle <- bundle(xgb_fit)Now, starting up a fresh R session and predicting on new data:
r(
function(model_bundle, new_data) {
library(bundle)
model_object <- unbundle(model_bundle)
predict(model_object, new_data)
},
args = list(
model_bundle = xgb_bundle,
new_data = x_test
)
)
#> Fiat X1-9 Porsche 914-2 Lotus Europa Ford Pantera L
#> 1.41180277 0.42267847 0.83762974 -0.10181840
#> Ferrari Dino Maserati Bora Volvo 142E
#> 0.09857675 -0.81635916 0.16201633Voilà! We hope bundles are helpful in making your modeling and deployment workflows a good bit smoother in the future.